Influenza Sequence Mapping Project
The aim of this project is to make information about the influenza virus available for exploration and discovery.
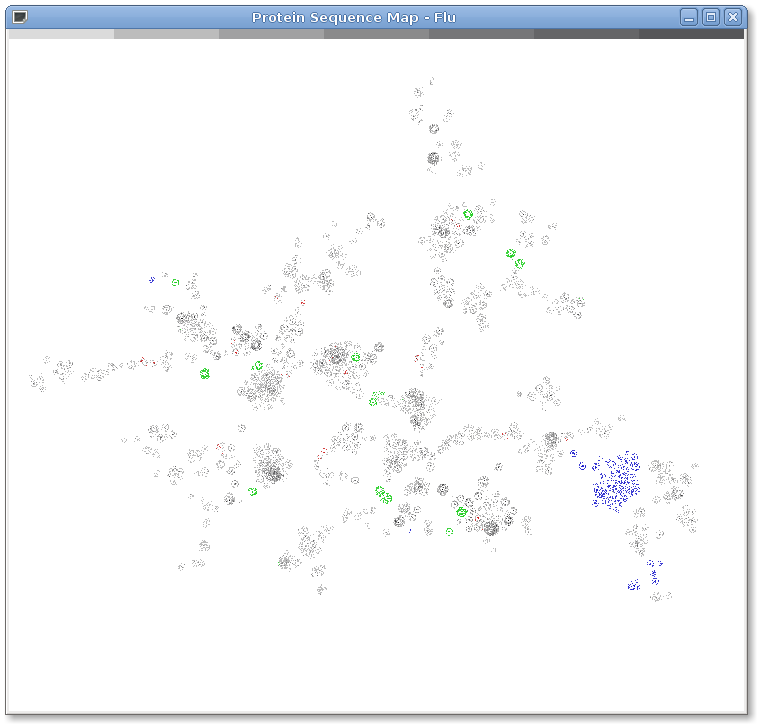
Figure 1: The genetic diversity of influenza. 114,996 influenza protein sequences from NCBI as of August 7, 2009. Protein sequences are represented as dots on the map. Position is determined by sequences similarity as calculated by BLASTP. Similar sequences appear closer to each other in the map. Layout is calculated by LGL. Sequences from the 2009 H1N1 Swine Flu pandemic are colored green. Sequences from the 1918 H1N1, 1957 H2N2 and 1968 H3N2 are colored red. Sequences that code for the PB1-F2 protein known to cause virulence in humans are colored blue. In this figure influenza is interpreted as a modular system with any given strain composed of approximately eight proteins from the display. The color bar at the top shows density with the lightest color indicating a single point and the darkest indicating seven or more overlapping points.
Resources
Discussion Board - An open board where anyone can discuss the project. A forum on analyses includes topics regarding interesting paths and connections made through the data. A forum on the visualization tool includes topics regarding the interactive tool for navigating the map and exploring the data.
Source Control Management System - A custom interactive visualization tool has been written to allow fast and efficient exploration of the map. This tool is implemented in C and OpenGL so that any available graphics hardware can be fully utilized to handle the large amount of data. The source code is stored in a git repository so that anyone can see the code and make contributions to the tool. This code is currently in development. It can be downloaded through the cgit interface available here. For now contributions of diffs and code changes should be emailed to the author for integration with the base.
Data - The data page documents the data collection and processing procedures.
Acknowledgments
This research was supported in part by the National Institutes of Health through resources provided by the National Resource for Biomedical Supercomputing (P41 RR06009), which is part of the Pittsburgh Supercomputing Center.

